A new report published on 2 November in the journal Science demonstrates how machine learning models based on large genomic and ecological datasets can be used to extract evolutionary signals imprinted in the virus sequence that offer information about the original hosts (1). In a perspective published on the same day in Science, Mark Woolhouse, a professor of infectious disease epidemiology from the University of Edinburgh, highlights that “if we do not know the reservoir host and/or vector, it is harder to identify individuals and populations at greatest risk of infection and to design an effective public health response” (2).
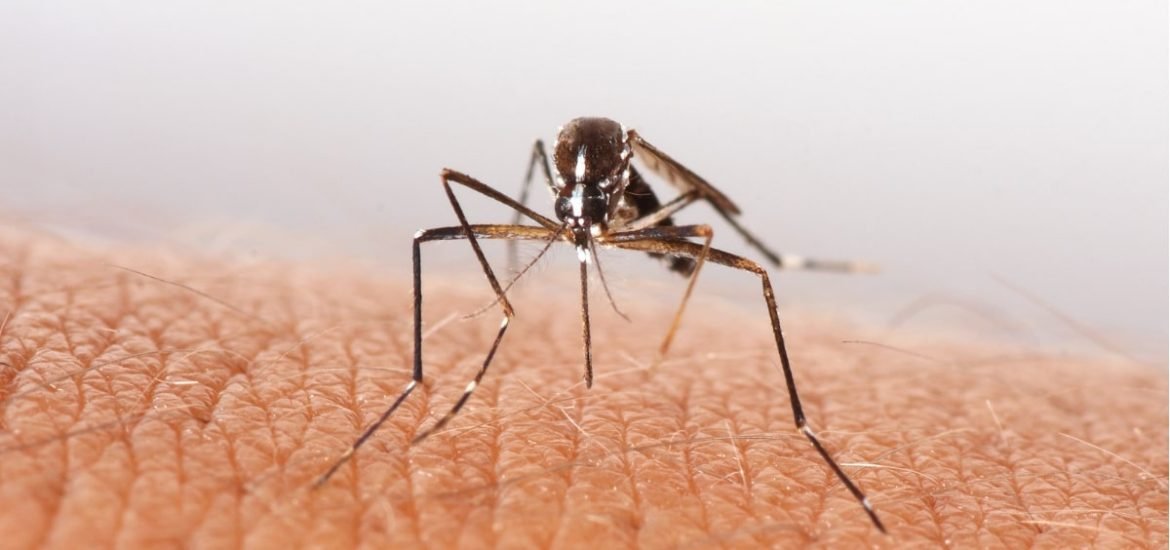
Preventing the further spread of viral infections—including Ebola, SARS, and Zika—hinges on the identification of the viral host. Normally, years of fieldwork and laboratory studies are required to identify the animal origins of RNA viruses. The lengthy discovery process comes with huge economic costs as well as costs in terms of human health and in some cases, fatalities. In this report, researchers from the University of Glasgow and the Moredun Research Institute demonstrate the ability of machine learning to predict the origins of some of the most human-infective single-stranded RNA viruses, including 69 viruses with previously unknown vectors.
As the accompanying perspective explains, “more than 200 species of RNA virus are known to be capable of infecting humans, and two or three new species are discovered every year.” And while little is often know about the biology of a new virus, its genome sequence can be obtained with relative ease. With access to this information, the researchers first built a model based on the idea that genetically similar viruses are more likely to have similar hosts and vectors. The algorithm was trained, optimised, and validated using more than 4000 genomic traits (such as amino acid bias or codon pair bias. The scientists then looked at genomic biases―the different combinations of letters that make up our DNA and the RNA of viruses―to decipher virus-host patterns.
The current model is a crude proof-of-concept. For known viruses, the general type of vector can be identified around 91 per cent of the time and the host reservoir type nearly 72 per cent of the time. But the report does illustrate the enormous potential for disease prevention. The proposed machine learning framework “leverages traits from individual viruses with network-derived information from their relatives” to predict a reservoir host and whether the transmission is through an arthropod vector, such as mosquitoes, fleas, ticks, and mites, and which one. As more new viruses are uncovered, the genomes and other details can be fed into the machine-learning model to improve its accuracy and specificity.
The authors suggest this method could be used for rapid assessment of emerging viruses for which we have no prior knowledge, referred to by the World Health Organization as “Disease X” scenarios. As Woolhouse writes, the technique “is a valuable step forward and hopefully presages further advances in our ability to extract information of public health value directly from virus genome sequences.”
(1) Babayan, S.A., Orton, R.J., and Streicker, D.G. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science (2018). DOI: 10.1126/science.aap9072
(2) Mark Woolhouse. Sources of human viruses. Science (2018). DOI: 10.1126/science.aav4265