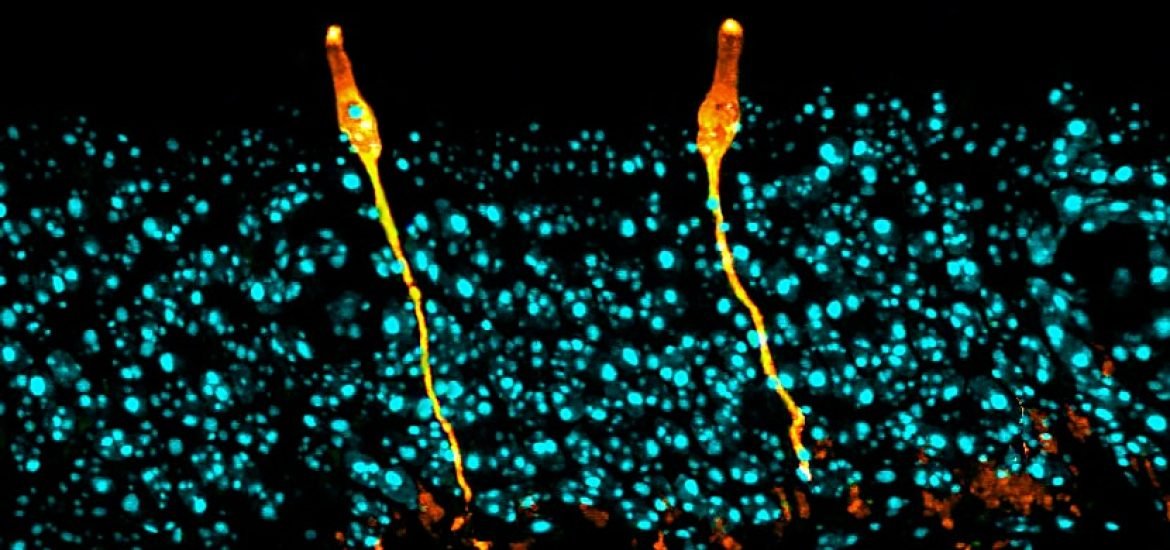
Two separate studies published on August 1 in Nature have identified the gene-expression patterns of tens of thousands of cells present in the lung and reported the existence of a previously unknown cell type in the human airway associated with cystic fibrosis ― the ‘pulmonary ionocyte.’ The two groups had set out to characterize the transcriptomes of tens of thousands of epithelial cells from the adult mouse trachea, on at a time, with the aim of exploring differences in the stem cell populations of the lung.
The new cell type is named after the ionocytes found in both fish gills and amphibian skin that have a similar gene expression pattern and play a role in balancing salt content. The cells contain the gene encoding for Foxi1, a transcription factor that influences ion transport.
In one of the studies, Dr. Aron Jaffe of Novartis and Dr. Allon Klein from Harvard joined forces to sequence the RNA of thousands of single human bronchial epithelial and mouse tracheal epithelial cells, thereby creating an atlas of the airway epithelium (1). This is what led to the discovery of the pulmonary ionocyte, in addition to characterizing the gene-expression patterns of other new, rare, or poorly understood cell types.
The study suggests this new cell type, discovered via the single-cell RNA-sequencing technique, may express the majority cystic fibrosis transmembrane conductance regulator (CFTR), the gene mutated in cystic fibrosis, despite making up only around 1 percent of airway cells. The chronic inherited disease for which there is still no cure is caused by the excessive production of dense mucus. The CTFR gene contains instructions for making the CFTR protein and alterations in this protein result in the buildup of mucus.
The other study, led by Jayaraj Rajagopal of Massachusetts General Hospital and Harvard University and Dr. Aviv Regev of the Massachusetts Institute of Technology (MIT) and also Harvard, revealed the variation in gene expression patterns of lung cells and discovered new structures in the lung as well as unfamiliar cell differentiation pathways (2). The study also confirmed the presence of the pulmonary ionocyte using single-cell RNA-sequencing and in vivo lineage tracing.
Single-cell sequencing technology is allowing us to map various cell types of different tissues and leading to new discoveries. The findings of these two studies have a number of important implications. The identification of this new rare cell type may lead to a better understanding of the cellular contribution to cystic fibrosis and of the disease itself, which could help in selecting suitable targets for future therapies to treat the disease.
Moreover, up to now, our knowledge of the expression of genes coding for transcription factors and specific cell-surface markers of the lung has been limited. Therefore, these new insights will have a significant impact on pulmonary research and expand our current understanding of lung biology and pulmonary diseases.
(1) Plasschaert L.W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature (2018). DOI: 10.1038/s41586-018-0394-6
(2) Montoro D.T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature (2018). DOI: 10.1038/s41586-018-0393-7
Image: Montoro et al. Nature (2018)